![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 002005_Nocton-Soy_GAGTGG_L003_R1_001.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 4000000 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 63 |
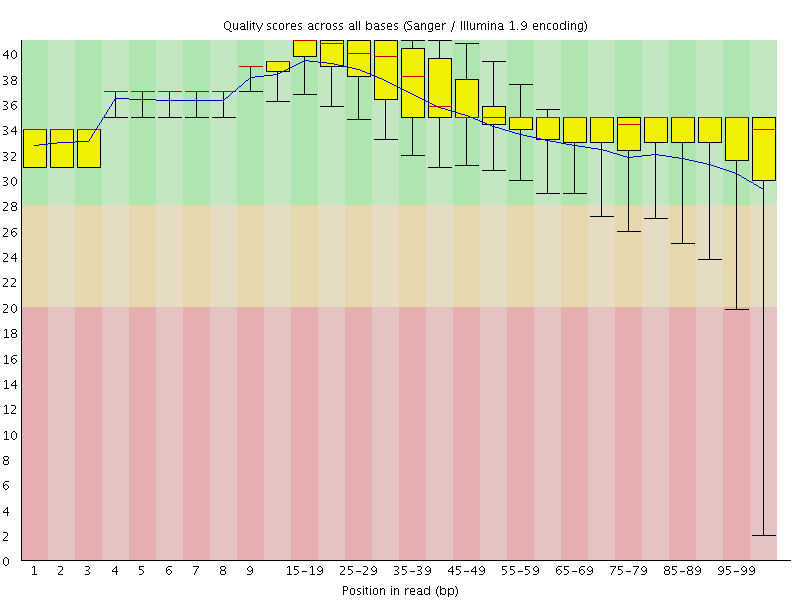
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
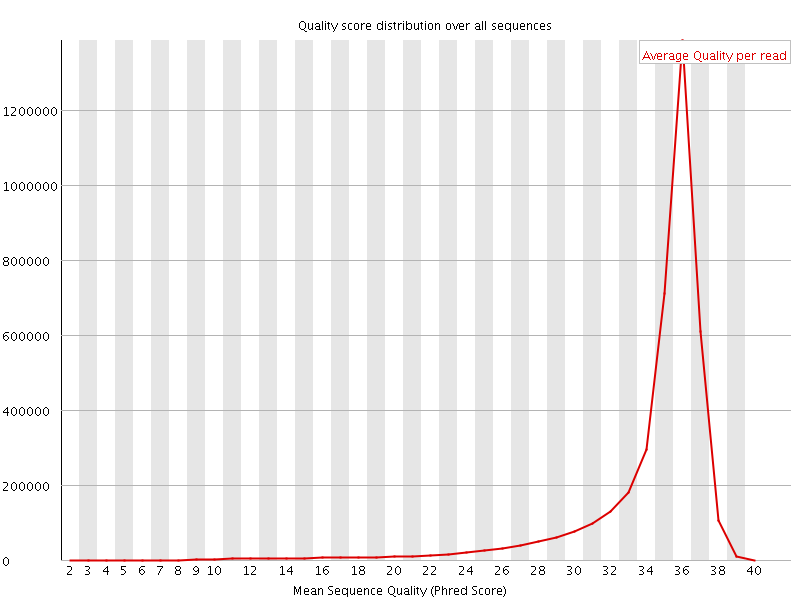
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
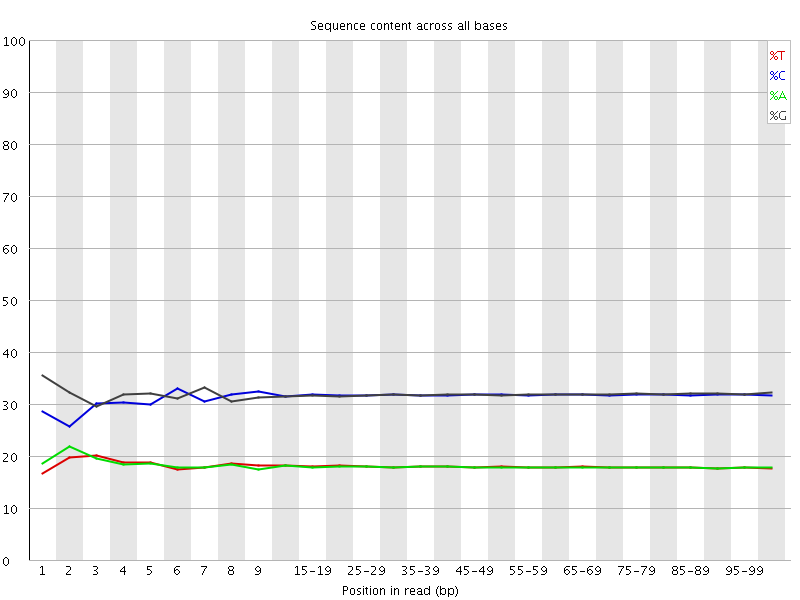
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
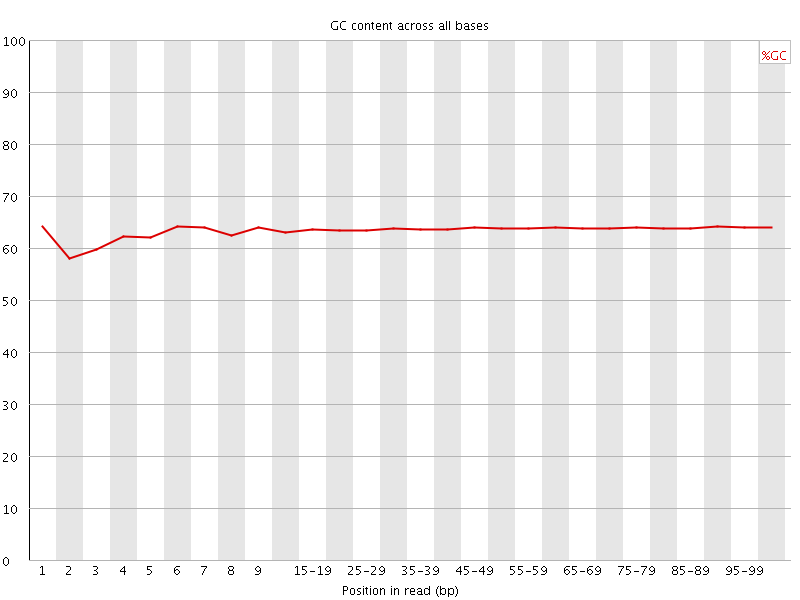
![[WARN]](Icons/warning.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
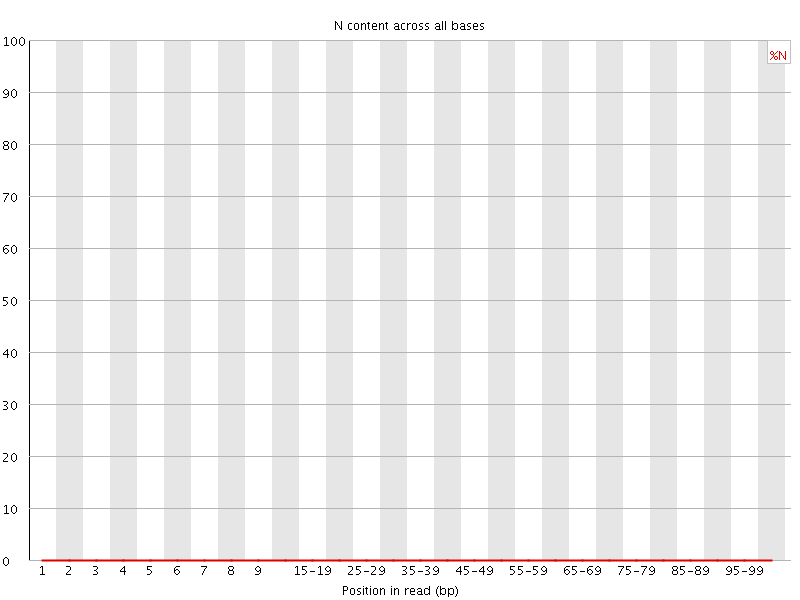
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
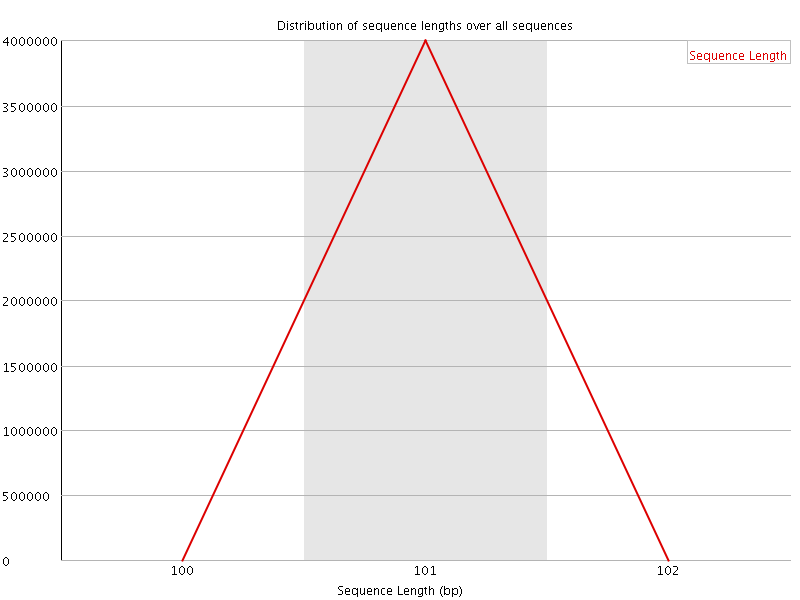
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
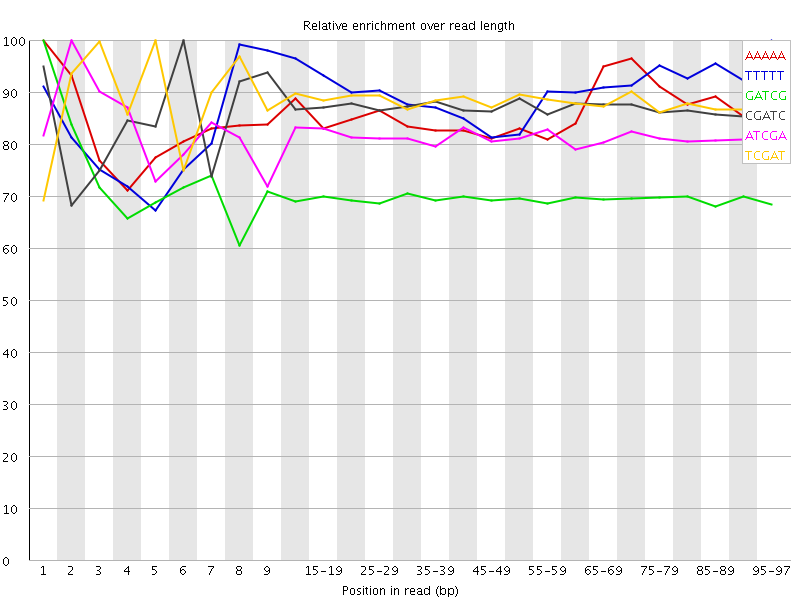
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 240635 | 3.1933126 | 3.7071114 | 1 |
| TTTTT | 236555 | 3.1316698 | 3.4883819 | 95-97 |
| GATCG | 1274930 | 3.0808613 | 4.4030895 | 1 |
| CGATC | 1263575 | 3.0720687 | 3.536716 | 6 |
| ATCGA | 710165 | 3.0333521 | 3.7218654 | 2 |
| TCGAT | 706970 | 3.0182605 | 3.4199114 | 5 |