![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 002005_Nocton-Soy_GAGTGG_L002_R2_008.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 4000000 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 63 |
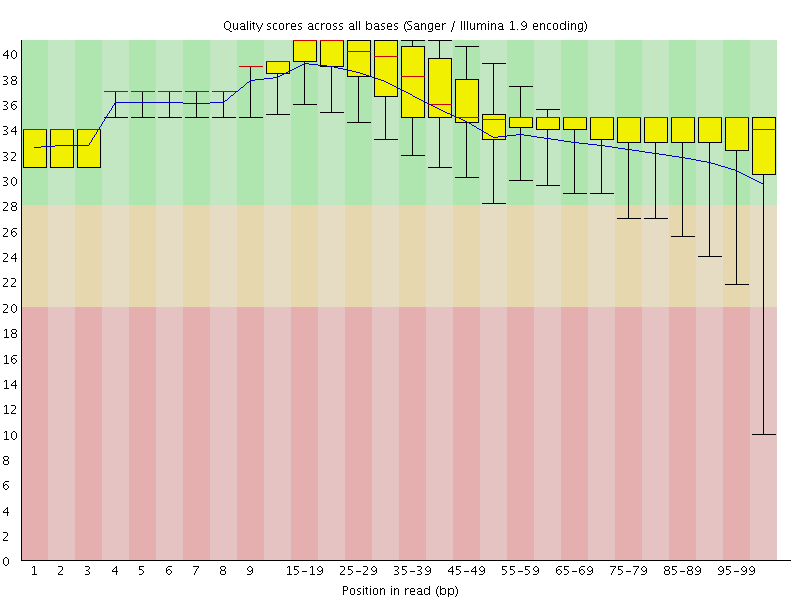
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
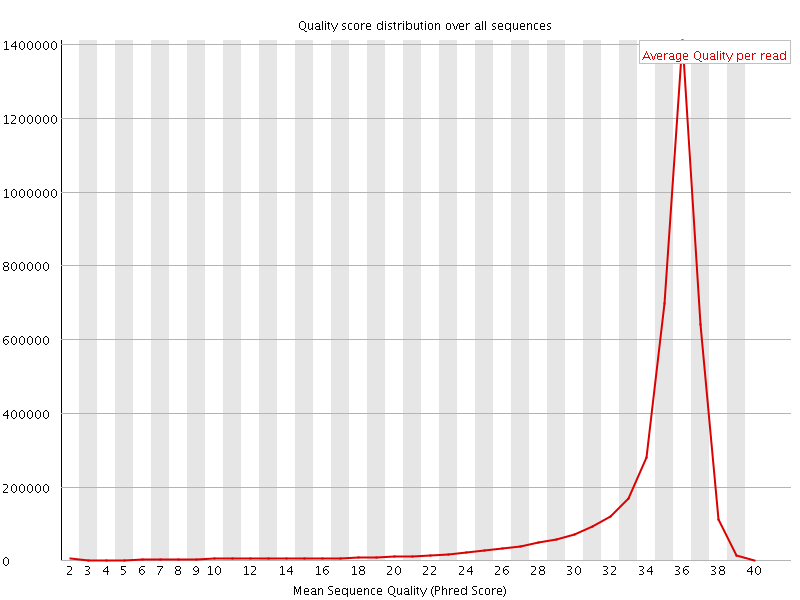
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
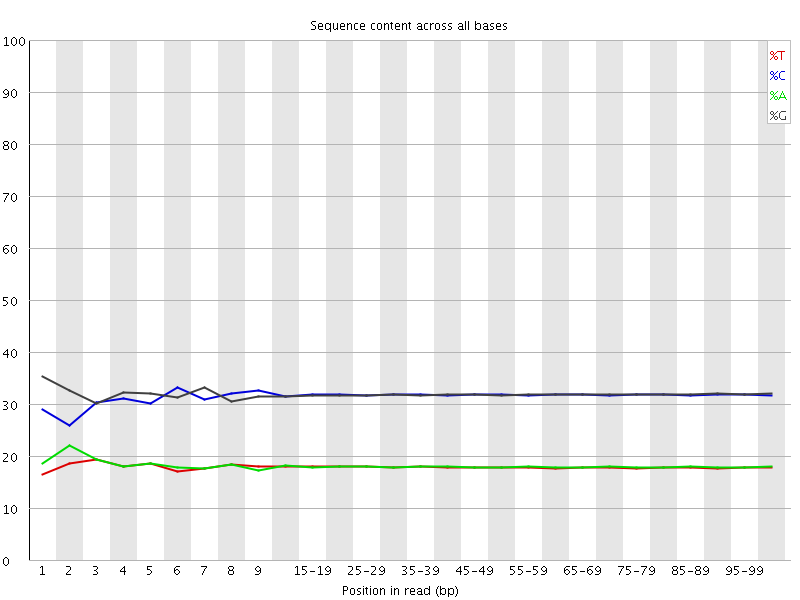
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
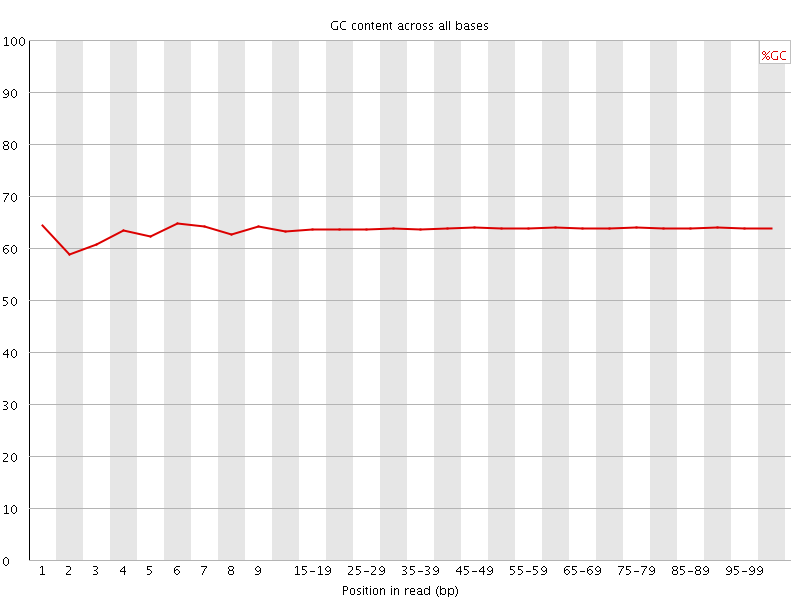
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
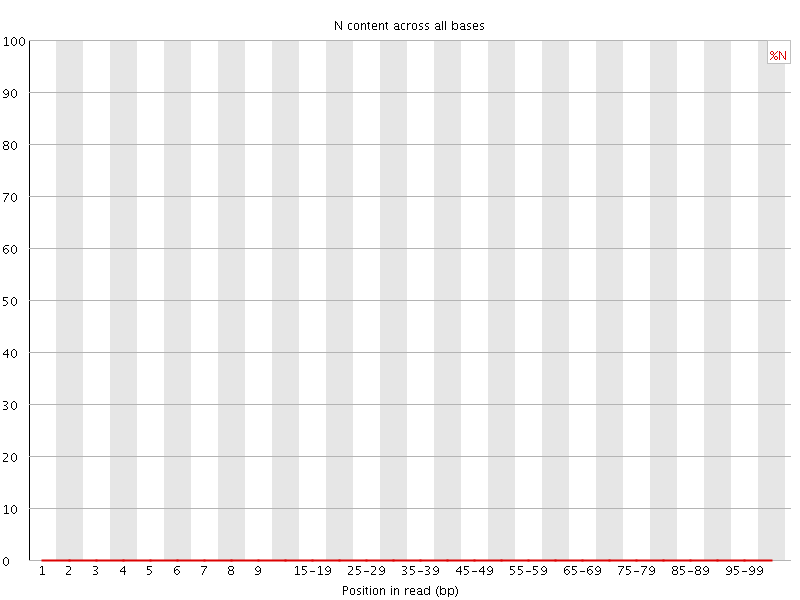
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
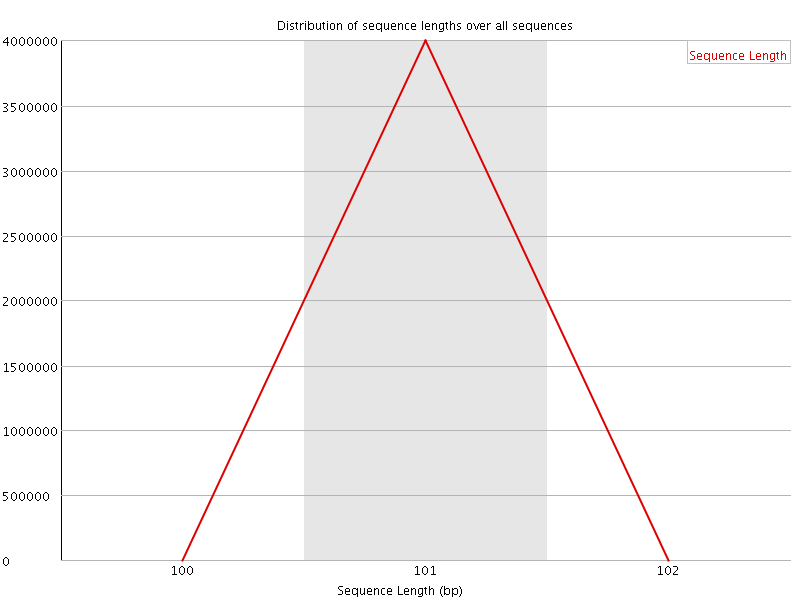
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
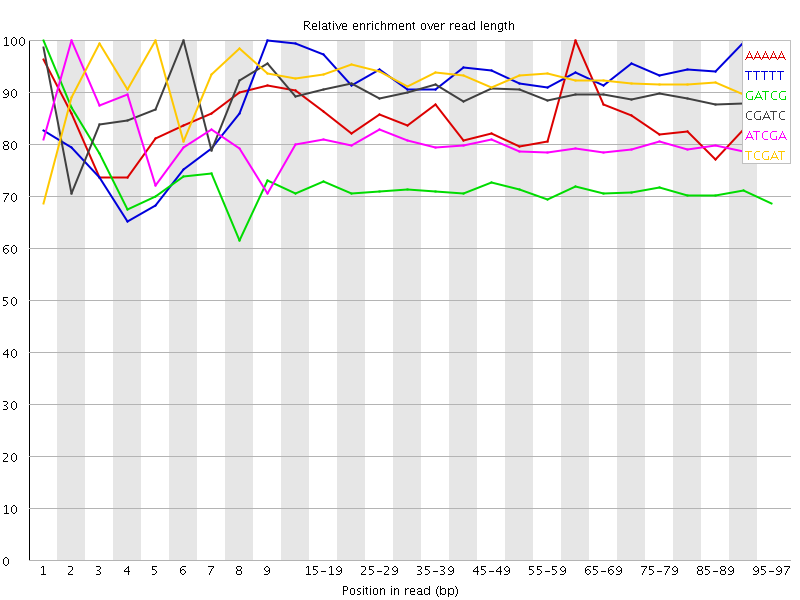
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 263660 | 3.476182 | 4.116747 | 60-64 |
| TTTTT | 238755 | 3.2355795 | 3.4920197 | 9 |
| GATCG | 1277870 | 3.0924423 | 4.32138 | 1 |
| CGATC | 1269715 | 3.0861714 | 3.4500246 | 6 |
| ATCGA | 710835 | 3.0368772 | 3.7912736 | 2 |
| TCGAT | 706445 | 3.034765 | 3.2853212 | 5 |