![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | 002005_Nocton-Soy_GAGTGG_L002_R2_003.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 4000000 |
| Filtered Sequences | 0 |
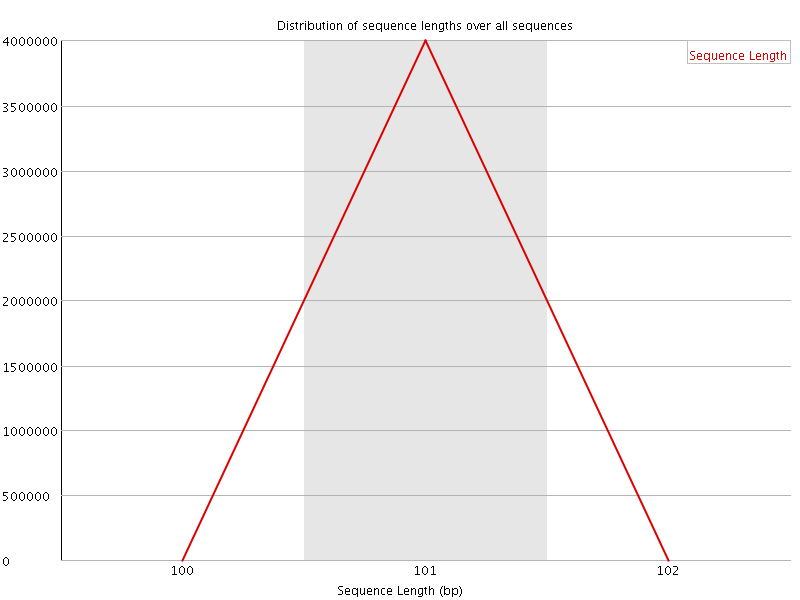
| Sequence length | 101 |
| %GC | 63 |
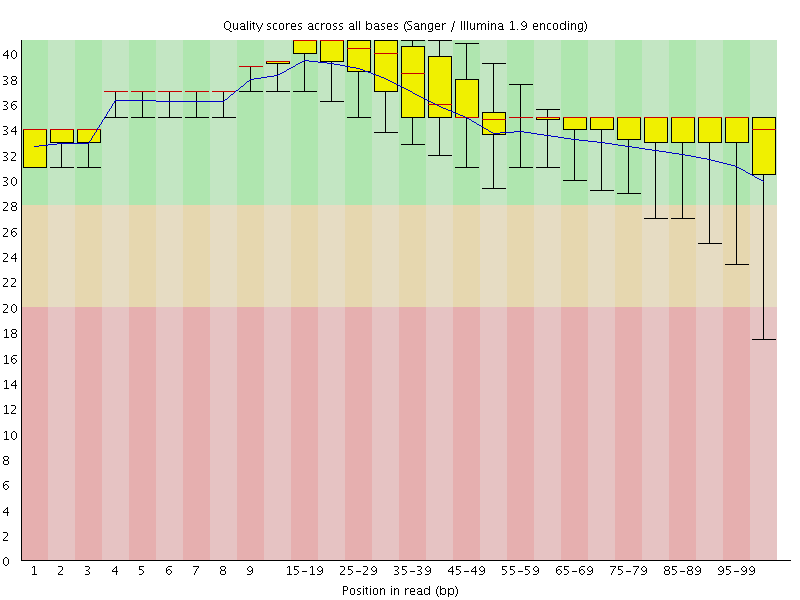
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
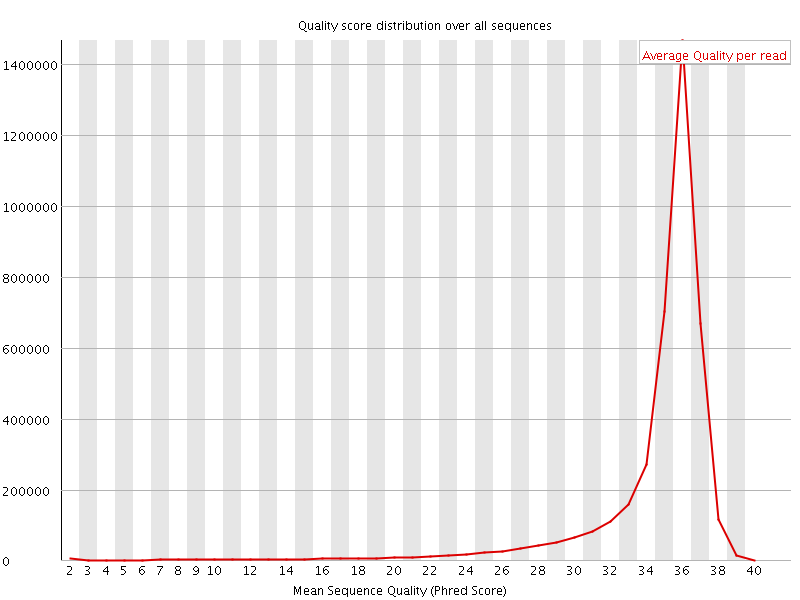
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
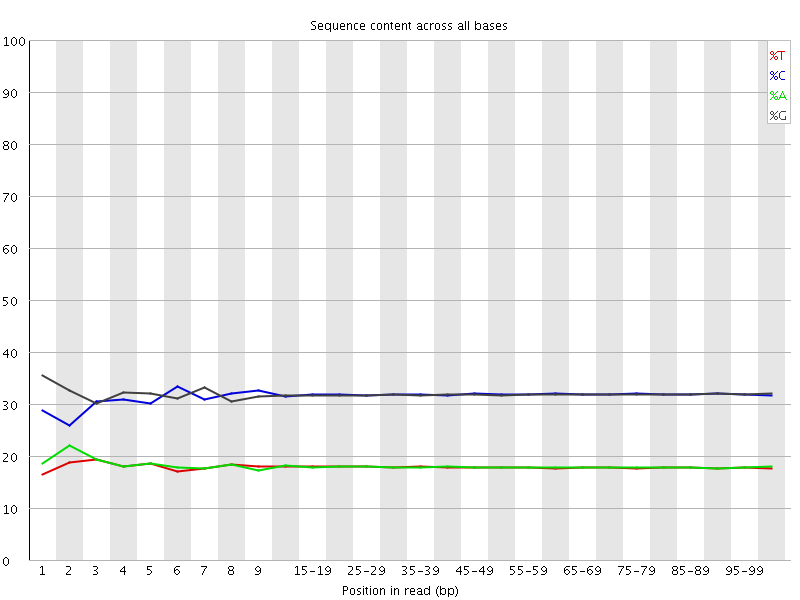
![[WARN]](Icons/warning.png) Per base sequence content
Per base sequence content
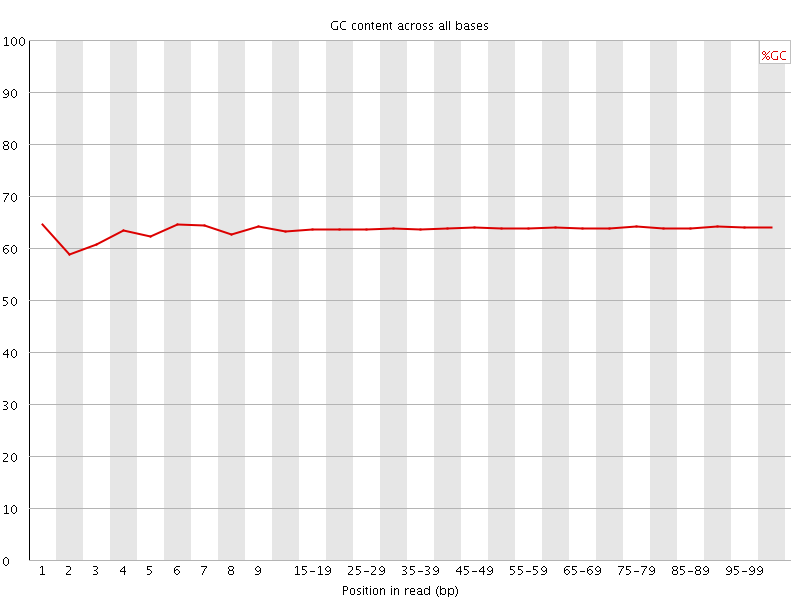
![[OK]](Icons/tick.png) Per base GC content
Per base GC content
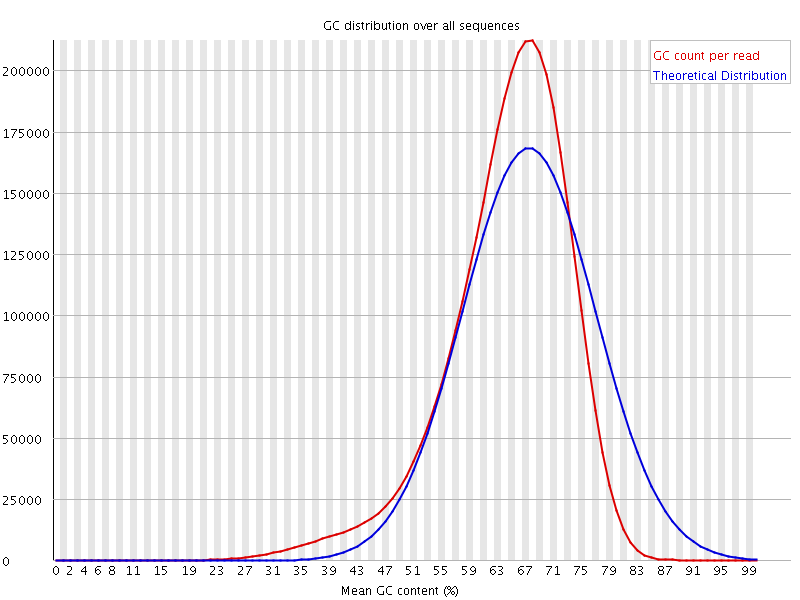
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
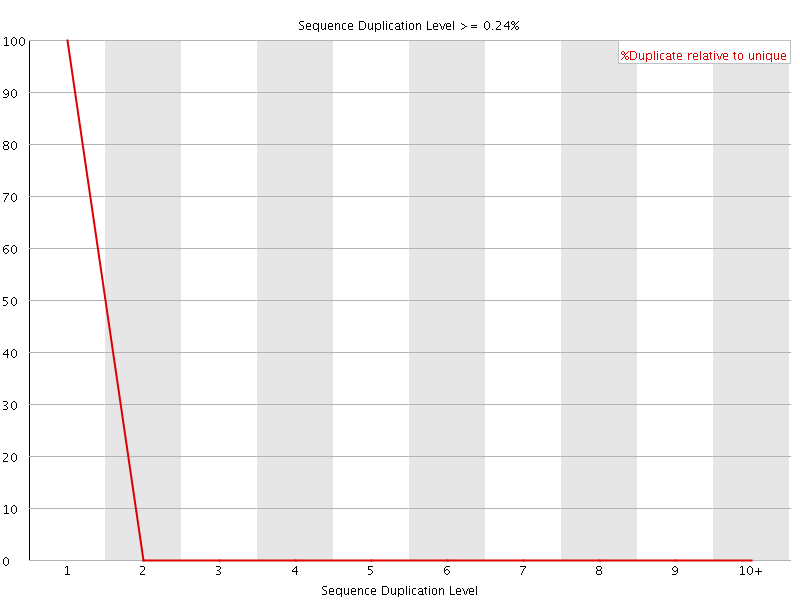
![[OK]](Icons/tick.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](Icons/tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
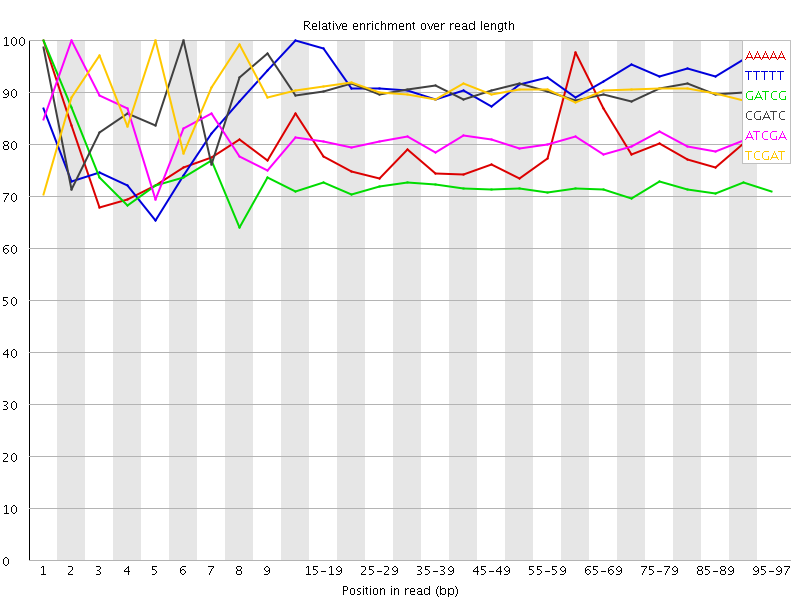
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| AAAAA | 249515 | 3.305965 | 4.187427 | 1 |
| TTTTT | 238170 | 3.2295785 | 3.5246031 | 10-14 |
| GATCG | 1275450 | 3.0879166 | 4.284147 | 1 |
| CGATC | 1267265 | 3.081872 | 3.4252007 | 6 |
| ATCGA | 712630 | 3.0496607 | 3.784747 | 2 |
| TCGAT | 705370 | 3.032605 | 3.3680158 | 5 |